Feature Engineering 1 - Missing values - Numerical
Missing Values - Feature Engineering
Lifecycle of Data Science projects
- Data Collection Strategy
- from company side
- 3rd party API’s
- Surveys
- Featue Engineering
- handling missing values
why are there missing values? - In case of surveys, people might have not filled the values - some input error while uploading numbers into system
-
In Data Science projects -> Data should be collected from multiple sources
-
Types of data that might be missings:-
- Continuous data
- Discrete continuous data (e.g age which will have integral value)
- Continuous data (e.g. height)
- categorial data
- Continuous data
What are different types of Missing data?
-
Missing completely at random (MCAR)
- a variable is missing completely at random (MCAR) if the probability of being missing is same for all the observations. When data is MCAR, there is absolutely no relationship between the data missing and any other values,observed or missing, within dataset. In other words, those missing data points are a random subset of the data. There is nothing systematic going on that makes some data more likely to be missing than other. if values for observation are missing completely at random, then disregarding those cases would not bias the inferences made.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
df=pd.read_csv('Datasets/Titanic/train.csv')
df.head()
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
df.isna().sum() #Number of missing values in each columns
PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 687
Embarked 2
dtype: int64
-
We can sense that there might be relationship between age and Cabin missing values as the person whose age is unknown cabin seems unknown for them, or person having null values in Age and Cabin may not have survived.
-
Missing values of Embarked seems MCAR.
df[df.Embarked.isna()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 61 | 62 | 1 | 1 | Icard, Miss. Amelie | female | 38.0 | 0 | 0 | 113572 | 80.0 | B28 | NaN |
| 829 | 830 | 1 | 1 | Stone, Mrs. George Nelson (Martha Evelyn) | female | 62.0 | 0 | 0 | 113572 | 80.0 | B28 | NaN |
-
Missing data not at random (MNAR) : Systematic missing value
- There is absolutely some relationship between the data missing and any other values, observed or missing, within the dataset.
df[df.Cabin.isna()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
| 5 | 6 | 0 | 3 | Moran, Mr. James | male | NaN | 0 | 0 | 330877 | 8.4583 | NaN | Q |
| 7 | 8 | 0 | 3 | Palsson, Master. Gosta Leonard | male | 2.0 | 3 | 1 | 349909 | 21.0750 | NaN | S |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 884 | 885 | 0 | 3 | Sutehall, Mr. Henry Jr | male | 25.0 | 0 | 0 | SOTON/OQ 392076 | 7.0500 | NaN | S |
| 885 | 886 | 0 | 3 | Rice, Mrs. William (Margaret Norton) | female | 39.0 | 0 | 5 | 382652 | 29.1250 | NaN | Q |
| 886 | 887 | 0 | 2 | Montvila, Rev. Juozas | male | 27.0 | 0 | 0 | 211536 | 13.0000 | NaN | S |
| 888 | 889 | 0 | 3 | Johnston, Miss. Catherine Helen "Carrie" | female | NaN | 1 | 2 | W./C. 6607 | 23.4500 | NaN | S |
| 890 | 891 | 0 | 3 | Dooley, Mr. Patrick | male | 32.0 | 0 | 0 | 370376 | 7.7500 | NaN | Q |
687 rows × 12 columns
#df['cabin_null']=df['Cabin'].apply(lambda x: 0 if x.isna() else 1)
df['cabin_null']=np.where(df.Cabin.isna(),1,0)
df[['Cabin','cabin_null']].head()
| Cabin | cabin_null | |
|---|---|---|
| 0 | NaN | 1 |
| 1 | C85 | 0 |
| 2 | NaN | 1 |
| 3 | C123 | 0 |
| 4 | NaN | 1 |
#find percentage of null values
df['cabin_null'].mean() #Alternate -> df.Cabin.isna().mean()
0.7710437710437711
df.columns
Index(['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp',
'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked', 'cabin_null'],
dtype='object')
Trying to figure out relationship of Cabin missing value with Survived column as we expect there should be less missing values for survived person
df.groupby('Survived').mean().cabin_null
Survived
0 0.876138
1 0.602339
Name: cabin_null, dtype: float64
-
Missing at Random (MAR)
- The probability of the missing values with respect to various features -> this will be almost same in the whole dataset.
- When the msising values have no relation with any other column but have relation with the column in which the value is missing
- e.g. Men hiding their salary, women hiding their age. (Kind of trend being analysed but not dependent to other variable)
All the techniques of handling missing values :-
- Mean/Median/Mode replacement
- Random Sample Imputation
- Capturing NAN values with a new feature
- End of Distribution imputation
- Arbitrary imputation
- Frequent categories imputation
1. Mean/Median/Mode imputation
When should we apply this?
- Mean/median imputation has the assumption that the data are missing completely at random (MCAR).
- Solve this by replacing the NAN with most frequent occurence of the variable.
- To overcome outlier we can use media/mode instead of mean.
df1=pd.read_csv('Datasets/Titanic/train.csv',usecols=['Age','Fare','Survived'])
df1.head()
| Survived | Age | Fare | |
|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 |
| 1 | 1 | 38.0 | 71.2833 |
| 2 | 1 | 26.0 | 7.9250 |
| 3 | 1 | 35.0 | 53.1000 |
| 4 | 0 | 35.0 | 8.0500 |
#Lets go and see the percentage of missing values
df1.isna().mean()
Survived 0.000000
Age 0.198653
Fare 0.000000
dtype: float64
def impute_nan(df,variable,method):
if method=='median':
value=df[variable].median()
elif method=='mean':
value=df[variable].mean()
elif method=='mode':
value=df[variable].mode()
df[variable+"_"+method]=df[variable].fillna(value)
impute_nan(df1,'Age','median')
impute_nan(df1,'Age','mode')
impute_nan(df1,'Age','mean')
df1[df1.Age.isna()]
| Survived | Age | Fare | Age_median | Age_mode | Age_mean | |
|---|---|---|---|---|---|---|
| 5 | 0 | NaN | 8.4583 | 28.0 | NaN | 29.699118 |
| 17 | 1 | NaN | 13.0000 | 28.0 | NaN | 29.699118 |
| 19 | 1 | NaN | 7.2250 | 28.0 | NaN | 29.699118 |
| 26 | 0 | NaN | 7.2250 | 28.0 | NaN | 29.699118 |
| 28 | 1 | NaN | 7.8792 | 28.0 | NaN | 29.699118 |
| ... | ... | ... | ... | ... | ... | ... |
| 859 | 0 | NaN | 7.2292 | 28.0 | NaN | 29.699118 |
| 863 | 0 | NaN | 69.5500 | 28.0 | NaN | 29.699118 |
| 868 | 0 | NaN | 9.5000 | 28.0 | NaN | 29.699118 |
| 878 | 0 | NaN | 7.8958 | 28.0 | NaN | 29.699118 |
| 888 | 0 | NaN | 23.4500 | 28.0 | NaN | 29.699118 |
177 rows × 6 columns
- Now we can check whether the standard deviation of age column changed if we compare the same with age_median
print('Age StdDev :-',df1.Age.std())
print('Age_median StdDev :-',df1.Age_median.std())
Age StdDev :- 14.526497332334044
Age_median StdDev :- 13.019696550973194
- Now we can check distributiojn of Age_median w.r.t. Age column using matplotlib
fig=plt.figure(figsize=(10,7))
ax=fig.add_subplot(111)
df1['Age'].plot(kind='kde',ax=ax)
df1.Age_median.plot(kind='kde',ax=ax,color='red')
lines,labels=ax.get_legend_handles_labels()
ax.legend(lines,labels,loc='best')
<matplotlib.legend.Legend at 0x7f98f26f7f10>
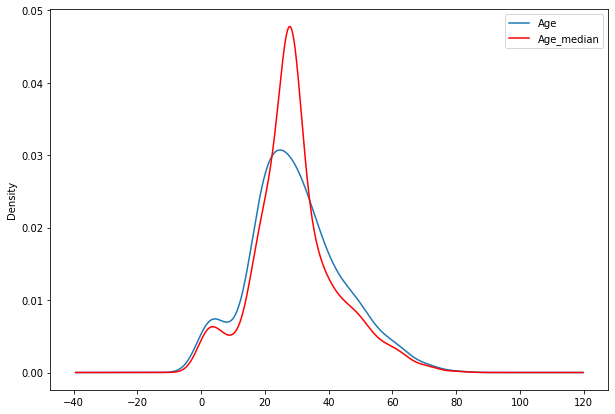
Advantages and disadvantages of Mean/Median imputations
Advantages :-
1. Easy to implement.
2. Robust to outliers (Median only).
3. Faster way to obtain complete datasets. #### Disadvantages :-
1. Change or distortion in the original variance/standard deviation.
2. It impacts correlation
2. Random Sample Imputation
Aim: Random sample imputation consists of taking random observation from the dataset and we use this observation to replace NAN value.
When shout it be used?
It assumes that data are missing completely at random (MCAR).
df2=pd.read_csv('Datasets/Titanic/train.csv',usecols=['Survived','Age','Fare'])
df2.head()
| Survived | Age | Fare | |
|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 |
| 1 | 1 | 38.0 | 71.2833 |
| 2 | 1 | 26.0 | 7.9250 |
| 3 | 1 | 35.0 | 53.1000 |
| 4 | 0 | 35.0 | 8.0500 |
df2.isnull().sum()
Survived 0
Age 177
Fare 0
dtype: int64
df2.isnull().mean()
Survived 0.000000
Age 0.198653
Fare 0.000000
dtype: float64
df['Age'].dropna().sample(df.Age.isna().sum(),random_state=0)
423 28.00
177 50.00
305 0.92
292 36.00
889 26.00
...
539 22.00
267 25.00
352 15.00
99 34.00
689 15.00
Name: Age, Length: 177, dtype: float64
def impute_nan_rand(df,variable,method):
if method!='random':
if method=='median':
value=df[variable].median()
elif method=='mean':
value=df[variable].mean()
elif method=='mode':
value=df[variable].mode()
df[variable+"_"+method]=df[variable].fillna(value)
else:
## Just to itroduce variable_random column which will be updated later
df[variable+"_random"]=df[variable]
## it will have the random sample to fill NAN value
random_sample=df['Age'].dropna().sample(df.Age.isna().sum(),random_state=0)
## pandas need to have same index in order to merge the datasets
random_sample.index=df[df[variable].isna()].index
#updating variable_random column with random sample for NAN values
df.loc[df[variable].isna(),variable+'_random']=random_sample
impute_nan_rand(df2,'Age','random')
df2
| Survived | Age | Fare | Age_random | |
|---|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 | 22.0 |
| 1 | 1 | 38.0 | 71.2833 | 38.0 |
| 2 | 1 | 26.0 | 7.9250 | 26.0 |
| 3 | 1 | 35.0 | 53.1000 | 35.0 |
| 4 | 0 | 35.0 | 8.0500 | 35.0 |
| ... | ... | ... | ... | ... |
| 886 | 0 | 27.0 | 13.0000 | 27.0 |
| 887 | 1 | 19.0 | 30.0000 | 19.0 |
| 888 | 0 | NaN | 23.4500 | 15.0 |
| 889 | 1 | 26.0 | 30.0000 | 26.0 |
| 890 | 0 | 32.0 | 7.7500 | 32.0 |
891 rows × 4 columns
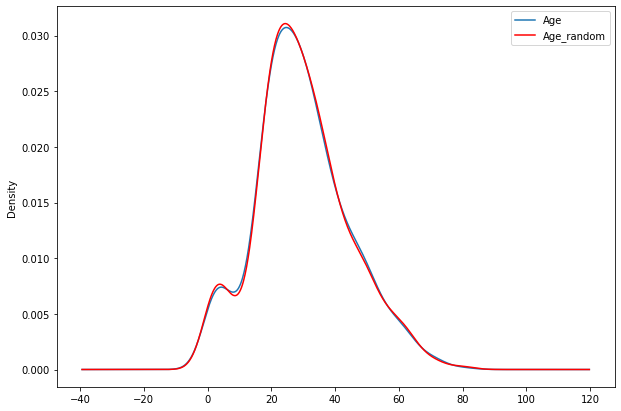
print('Age StdDev :-',df2.Age.std())
print('Age_random StdDev :-',df2.Age_random.std())
Age StdDev :- 14.526497332334044
Age_random StdDev :- 14.5636540895687
fig=plt.figure(figsize=(10,7))
ax=fig.add_subplot(111)
df2['Age'].plot(kind='kde',ax=ax)
df2.Age_random.plot(kind='kde',ax=ax,color='red')
lines,labels=ax.get_legend_handles_labels()
ax.legend(lines,labels,loc='best')
<matplotlib.legend.Legend at 0x7f98f3f8da10>
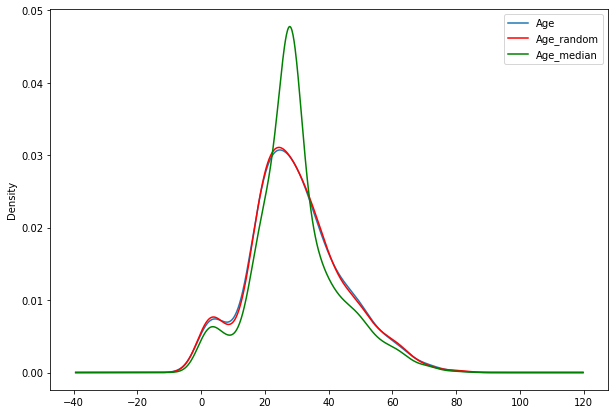
impute_nan_rand(df2,'Age','median')
fig=plt.figure(figsize=(10,7))
ax=fig.add_subplot(111)
df2['Age'].plot(kind='kde',ax=ax)
df2.Age_random.plot(kind='kde',ax=ax,color='red')
df2.Age_median.plot(kind='kde',ax=ax,color='green')
lines,labels=ax.get_legend_handles_labels()
ax.legend(lines,labels,loc='best')
<matplotlib.legend.Legend at 0x7f98f410bd50>
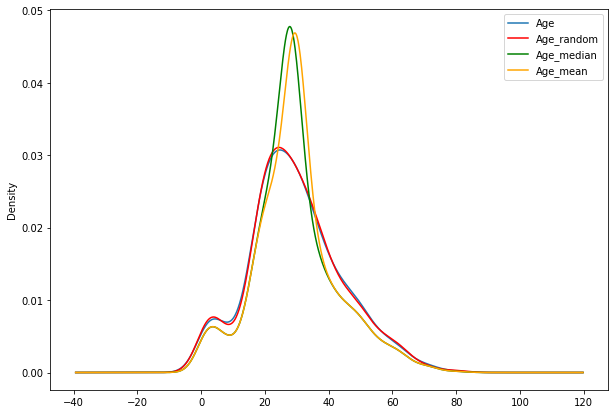
impute_nan_rand(df2,'Age','mean')
fig=plt.figure(figsize=(10,7))
ax=fig.add_subplot(111)
df2['Age'].plot(kind='kde',ax=ax)
df2.Age_random.plot(kind='kde',ax=ax,color='red')
df2.Age_median.plot(kind='kde',ax=ax,color='green')
df2.Age_mean.plot(kind='kde',ax=ax,color='orange')
lines,labels=ax.get_legend_handles_labels()
ax.legend(lines,labels,loc='best')
<matplotlib.legend.Legend at 0x7f98f40e2310>
Advantages
- Easy to implement
- Less distortion in variance or standard deviation
Disadvantages
- In every situation randomness won’t work
3. Capturing NAN values with a new feature
It works well if the data are missing completely not at random (MNAR).
df3=pd.read_csv('Datasets/Titanic/train.csv',usecols=['Survived','Age','Fare'])
df3.head()
| Survived | Age | Fare | |
|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 |
| 1 | 1 | 38.0 | 71.2833 |
| 2 | 1 | 26.0 | 7.9250 |
| 3 | 1 | 35.0 | 53.1000 |
| 4 | 0 | 35.0 | 8.0500 |
#Just to flag the missing value, from this we can identify which row has been imputed from missing value
df3['Age_NAN']=np.where(df3.Age.isna(),1,0)
df3.Age_NAN
0 0
1 0
2 0
3 0
4 0
..
886 0
887 0
888 1
889 0
890 0
Name: Age_NAN, Length: 891, dtype: int64
df3
| Survived | Age | Fare | Age_NAN | |
|---|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 | 0 |
| 1 | 1 | 38.0 | 71.2833 | 0 |
| 2 | 1 | 26.0 | 7.9250 | 0 |
| 3 | 1 | 35.0 | 53.1000 | 0 |
| 4 | 0 | 35.0 | 8.0500 | 0 |
| ... | ... | ... | ... | ... |
| 886 | 0 | 27.0 | 13.0000 | 0 |
| 887 | 1 | 19.0 | 30.0000 | 0 |
| 888 | 0 | NaN | 23.4500 | 1 |
| 889 | 1 | 26.0 | 30.0000 | 0 |
| 890 | 0 | 32.0 | 7.7500 | 0 |
891 rows × 4 columns
df3['Age'].fillna(df3.Age.median(),inplace=True)
df3
| Survived | Age | Fare | Age_NAN | |
|---|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 | 0 |
| 1 | 1 | 38.0 | 71.2833 | 0 |
| 2 | 1 | 26.0 | 7.9250 | 0 |
| 3 | 1 | 35.0 | 53.1000 | 0 |
| 4 | 0 | 35.0 | 8.0500 | 0 |
| ... | ... | ... | ... | ... |
| 886 | 0 | 27.0 | 13.0000 | 0 |
| 887 | 1 | 19.0 | 30.0000 | 0 |
| 888 | 0 | 28.0 | 23.4500 | 1 |
| 889 | 1 | 26.0 | 30.0000 | 0 |
| 890 | 0 | 32.0 | 7.7500 | 0 |
891 rows × 4 columns
Advantages
- Easy to implement
- Captures the importance of missing values. (Age_NAN column giving importance to missing value as it is 1 for NAN cases)
Disadvantages
- Creating additional features, as number of features which have missing values increases we need to create same number of additional features. (Curse of Dimensionality)
4. End of Distribution imputation
If there is suspicion that the missing value is not at random then capturing that information is important. In this scenario, one would want to replace missing data with values that are at the tails of the distribution of the variable.
df4=pd.read_csv('Datasets/Titanic/train.csv',usecols=['Survived','Age','Fare'])
df4.head()
| Survived | Age | Fare | |
|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 |
| 1 | 1 | 38.0 | 71.2833 |
| 2 | 1 | 26.0 | 7.9250 |
| 3 | 1 | 35.0 | 53.1000 |
| 4 | 0 | 35.0 | 8.0500 |
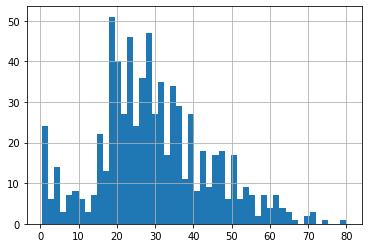
df4.Age.hist(bins=50)
<matplotlib.axes._subplots.AxesSubplot at 0x7f98f3887750>
#Mean
df4.Age.mean()
29.69911764705882
#Standard deviation
df4.Age.std()
14.526497332334044
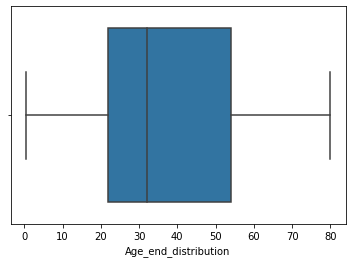
sns.boxplot('Age',data=df4)
<matplotlib.axes._subplots.AxesSubplot at 0x7f98f4e01ed0>
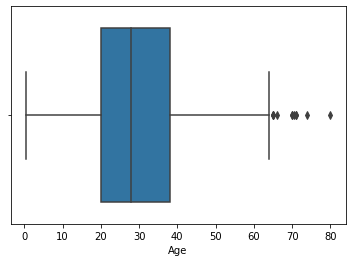
- In this dataset we have only right side outliers, therefore we will impute the missing values from roght extremem values
#Picking values from right end of distribution -> mean+3*standard_deviation
df4.Age.mean()+3*df4.Age.std()
73.27860964406095
def impute_nan_extreme(df,variable,method):
if method=='random':
## Just to itroduce variable_random column which will be updated later
df[variable+"_random"]=df[variable]
## it will have the random sample to fill NAN value
random_sample=df['Age'].dropna().sample(df.Age.isna().sum(),random_state=0)
## pandas need to have same index in order to merge the datasets
random_sample.index=df[df[variable].isna()].index
#updating variable_random column with random sample for NAN values
df.loc[df[variable].isna(),variable+'_random']=random_sample
elif method=='extreme':
#getting the right extreme value (mean+3*std_dev)
value=df[variable].mean()+3*df[variable].std()
df[variable+"_end_distribution"]=df[variable].fillna(value)
else:
if method=='median':
value=df[variable].median()
elif method=='mean':
value=df[variable].mean()
elif method=='mode':
value=df[variable].mode()
df[variable+"_"+method]=df[variable].fillna(value)
impute_nan_extreme(df4,'Age','extreme')
df4
| Survived | Age | Fare | Age_end_distribution | |
|---|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 | 22.00000 |
| 1 | 1 | 38.0 | 71.2833 | 38.00000 |
| 2 | 1 | 26.0 | 7.9250 | 26.00000 |
| 3 | 1 | 35.0 | 53.1000 | 35.00000 |
| 4 | 0 | 35.0 | 8.0500 | 35.00000 |
| ... | ... | ... | ... | ... |
| 886 | 0 | 27.0 | 13.0000 | 27.00000 |
| 887 | 1 | 19.0 | 30.0000 | 19.00000 |
| 888 | 0 | NaN | 23.4500 | 73.27861 |
| 889 | 1 | 26.0 | 30.0000 | 26.00000 |
| 890 | 0 | 32.0 | 7.7500 | 32.00000 |
891 rows × 4 columns
impute_nan_extreme(df4,'Age','median')
df4
| Survived | Age | Fare | Age_end_distribution | Age_median | |
|---|---|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 | 22.00000 | 22.0 |
| 1 | 1 | 38.0 | 71.2833 | 38.00000 | 38.0 |
| 2 | 1 | 26.0 | 7.9250 | 26.00000 | 26.0 |
| 3 | 1 | 35.0 | 53.1000 | 35.00000 | 35.0 |
| 4 | 0 | 35.0 | 8.0500 | 35.00000 | 35.0 |
| ... | ... | ... | ... | ... | ... |
| 886 | 0 | 27.0 | 13.0000 | 27.00000 | 27.0 |
| 887 | 1 | 19.0 | 30.0000 | 19.00000 | 19.0 |
| 888 | 0 | NaN | 23.4500 | 73.27861 | 28.0 |
| 889 | 1 | 26.0 | 30.0000 | 26.00000 | 26.0 |
| 890 | 0 | 32.0 | 7.7500 | 32.00000 | 32.0 |
891 rows × 5 columns
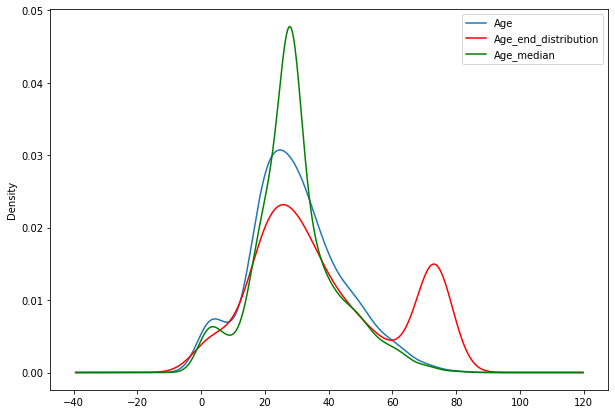
fig=plt.figure(figsize=(10,7))
ax=fig.add_subplot(111)
df4['Age'].plot(kind='kde',ax=ax)
df4.Age_end_distribution.plot(kind='kde',ax=ax,color='red')
df4.Age_median.plot(kind='kde',ax=ax,color='green')
lines,labels=ax.get_legend_handles_labels()
ax.legend(lines,labels,loc='best')
<matplotlib.legend.Legend at 0x7f98f4ee6350>
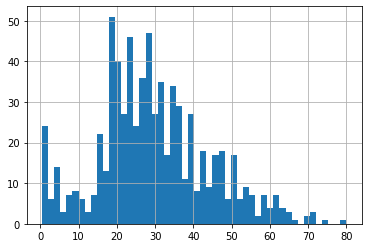
df4.Age.hist(bins=50)
<matplotlib.axes._subplots.AxesSubplot at 0x7f98f5ebfc10>
df4.Age_median.hist(bins=50)
<matplotlib.axes._subplots.AxesSubplot at 0x7f98f58e5f50>
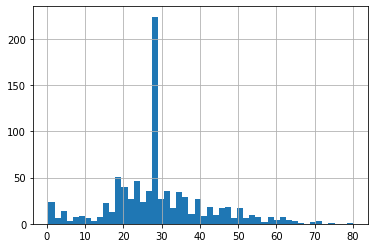
df4.Age_end_distribution.hist(bins=50)
<matplotlib.axes._subplots.AxesSubplot at 0x7f98f5a84f10>
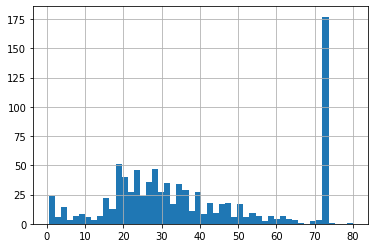
sns.boxplot('Age_end_distribution',data=df4)
<matplotlib.axes._subplots.AxesSubplot at 0x7f98f6042190>
- we can see that there is no more outliers after end of distribution imputation.
- We can observe skewness in the distribution after imputation.
Advantages
- Easy to implement
- Captures the importance of missing values if there is one
Disadvantages
- Distorts the original distribution of the variable
- If missingness is not important, it may mask the predictive power of the original variable by distorting its distribution.
- If number of NAN is big, it will mask true outliers in the distribution
- If the number of NAN is small, the replaced NAN may be considered as outlier and pre-processed in subsequent steps of feature engineering
Note
- if we are working in real world environment only one type of feature engineering will not be sufficient, we need to show all the observation/scenarios to the stakeholders.
5. Arbitrary imputation
- It consists of replacing NAN by an arbitrary value
- This technique was derived from kaggle competetions
df5=pd.read_csv('Datasets/Titanic/train.csv',usecols=['Survived','Age','Fare'])
df5.head()
| Survived | Age | Fare | |
|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 |
| 1 | 1 | 38.0 | 71.2833 |
| 2 | 1 | 26.0 | 7.9250 |
| 3 | 1 | 35.0 | 53.1000 |
| 4 | 0 | 35.0 | 8.0500 |
df5.Age.hist(bins=50)
<matplotlib.axes._subplots.AxesSubplot at 0x7f98f63dc250>
Arbitrary values :-
- It should not be the more frequently present in data
- we can either use right end (or > right end) or left end (or < left end)
def impute_arbitrary(df,variable):
df[variable+"_rightend"]=df[variable].fillna(df[variable].max()+20)
df[variable+"_leftend"]=df[variable].fillna(df[variable].min())
impute_arbitrary(df5,'Age')
df5
| Survived | Age | Fare | Age_rightend | Age_leftend | |
|---|---|---|---|---|---|
| 0 | 0 | 22.0 | 7.2500 | 22.0 | 22.00 |
| 1 | 1 | 38.0 | 71.2833 | 38.0 | 38.00 |
| 2 | 1 | 26.0 | 7.9250 | 26.0 | 26.00 |
| 3 | 1 | 35.0 | 53.1000 | 35.0 | 35.00 |
| 4 | 0 | 35.0 | 8.0500 | 35.0 | 35.00 |
| ... | ... | ... | ... | ... | ... |
| 886 | 0 | 27.0 | 13.0000 | 27.0 | 27.00 |
| 887 | 1 | 19.0 | 30.0000 | 19.0 | 19.00 |
| 888 | 0 | NaN | 23.4500 | 100.0 | 0.42 |
| 889 | 1 | 26.0 | 30.0000 | 26.0 | 26.00 |
| 890 | 0 | 32.0 | 7.7500 | 32.0 | 32.00 |
891 rows × 5 columns
Advantages
- Easy to implement
- Captures the importance of missing values if there is one
Disadvantages
- Distorts the original distribution of the variable
- If missingness is not important, it may mask the predictive power of the original variable by distorting its distribution.
- Hard to decide which value to use

